How PharmCAT Works
PharmCAT (Pharmacogenomics Clinical Annotation Tool) is a bioinformatics tool that analyzes genetic variants to predict drug response and tailor medical treatment to an individual patient’s genetic profile. It does this in two phases:
- Processes VCF files from next generation sequencing (NGS) or genotyping methods and identifies pharmacogenomic (PGx) genotypes and infers haplotypes, typically called star alleles.
- Uses the pharmacogene diplotypes (combination of maternal and paternal star alleles) to predict PGx phenotypes and reports the corresponding drug-prescribing recommendations from CPIC guidelines, PharmGKB-annotated DPWG guidelines and PharmGKB-annotated FDA-approved drug labels.
The PharmCAT pipeline can be visualized as such:
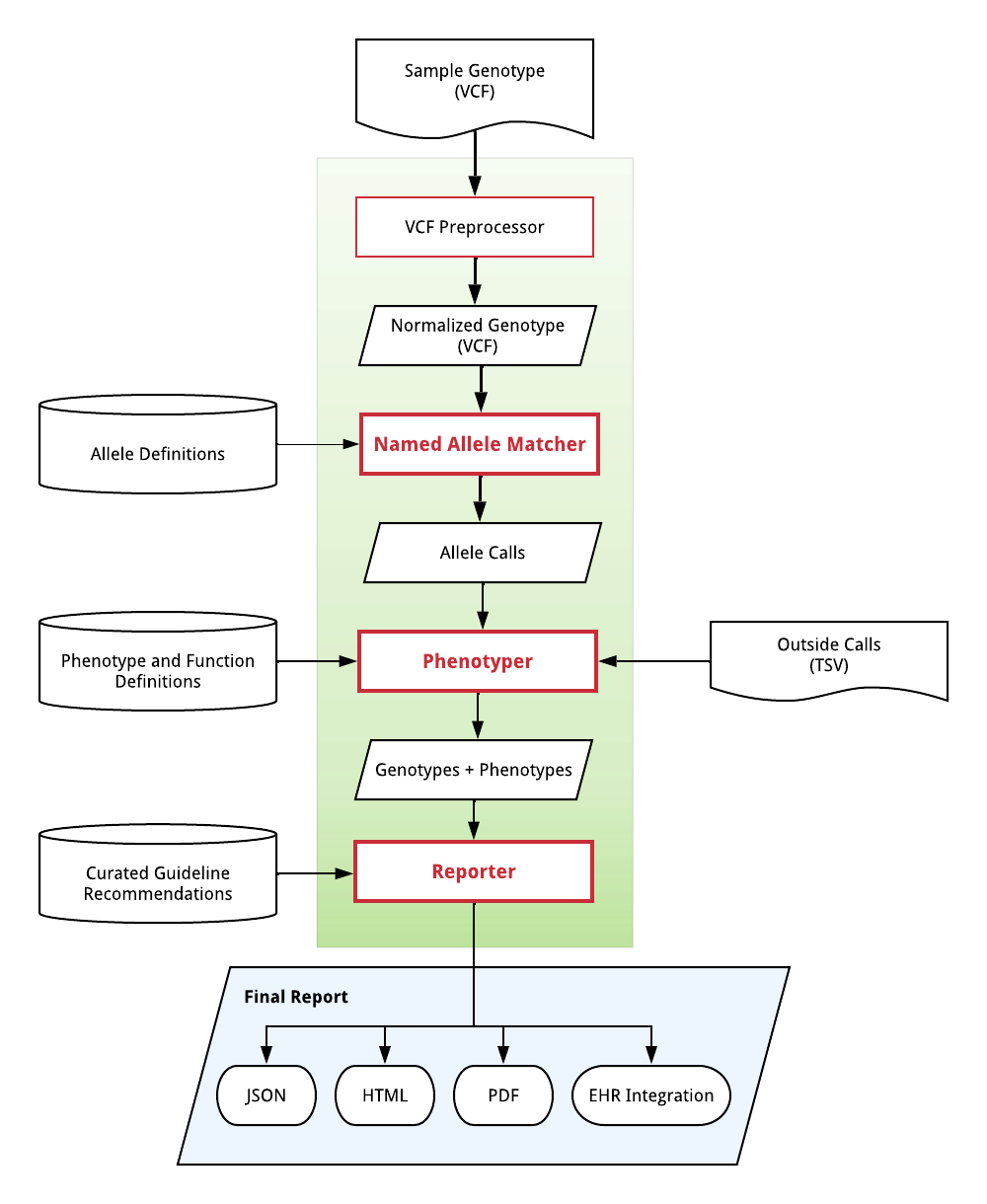
Phase 1 - Determining Alleles
The VCF Preprocessor module is responsible for normalizing the input VCF. See VCF Preprocessor for details.
The Named Allele Matcher module is responsible for calling diplotypes for a sample based on a VCF file. See:
- Named Allele Matcher 101 for an overview
- Named Allele Matcher 201 for details
Special cases and exceptions to how alleles are called are cataloged in Gene Definition Exceptions. For details on calling CYP2D6, please see Calling CYP2D6.
Phase 2 - Matching Recommendations
The Phenotyper module is responsible for translating diplotypes and other genotype information into gene-specific phenotypes. For example, CYP2C19 *2/*4 is a poor metabolizer.
The Reporter module is responsible for generating a report with genotype-specific expert-reviewed drug prescribing recommendations for clinical decision support.
See Matching Recommendations for details on how the Phenotyper and Reporter works.